Browse by Organisms Motifs
| M. musculus Motifs of MC08 group | ||||
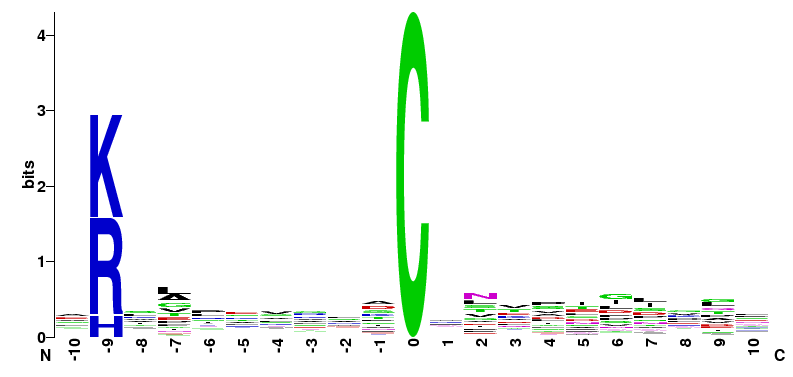 |
| No. | SwissProt ID | Position | S-nitrosylated Peptide | PubMed ID |
| 1 | 2AAA_MOUSE | 390 | VRLNIISNLD C VNEVIGIRQL | 19101475 |
| 2 | 2AAA_MOUSE | 390 | VRLNIISNLD C VNEVIGIRQL | 19483679 |
| 3 | 2AAA_MOUSE | 390 | VRLNIISNLD C VNEVIGIRQL | 21278135 |
| 4 | 3HIDH_MOUSE | 250 | AKILNMSSGR C WSSDTYNPVP | 21278135 |
| 5 | 3HIDH_MOUSE | 250 | AKILNMSSGR C WSSDTYNPVP | 22865876 |
| 6 | ABLM1_MOUSE | 314 | YHPSCARCSR C NQMFTEGEEM | 22865876 |
| 7 | ACAD9_MOUSE | 275 | DKLGIRGSNT C EVHFENTRVP | 21278135 |
| 8 | ACADL_MOUSE | 342 | HKLAELKTHI C VTRAFVDSCL | 22178444 |
| 9 | ACADL_MOUSE | 342 | HKLAELKTHI C VTRAFVDSCL | 21278135 |
| 10 | ACADL_MOUSE | 342 | HKLAELKTHI C VTRAFVDSCL | 20837516 |
| 11 | ACADL_MOUSE | 342 | HKLAELKTHI C VTRAFVDSCL | 22865876 |
| 12 | ACDSB_MOUSE | 261 | NKMGIRASST C QLTFENVKVP | 21278135 |
| 13 | ACLY_MOUSE | 835 | RKPASFMTSI C DERGQELIYA | 22178444 |
| 14 | ACLY_MOUSE | 835 | RKPASFMTSI C DERGQELIYA | 20837516 |
| 15 | ACON_MOUSE | 410 | AKQALAHGLK C KSQFTITPGS | 21278135 |
| 16 | ACON_MOUSE | 410 | AKQALAHGLK C KSQFTITPGS | 20837516 |
| 17 | ACS2L_MOUSE | 41 | SRSTATRLPG C VPAAAQPGSY | 21278135 |
| 18 | ACS2L_MOUSE | 41 | SRSTATRLPG C VPAAAQPGSY | 22865876 |
| 19 | ACTN1_MOUSE | 332 | LHKPPKVQEK C QLEINFNTLQ | 20925432 |
| 20 | ACTN2_MOUSE | 339 | KHKPPKVQEK C QLEINFNTLQ | 22865876 |
| 21 | ACTN4_MOUSE | 352 | VHKPPKVQEK C QLEINFNTLQ | 20925432 |
| 22 | AINX_MOUSE | 54 | SRSNVASTAA C SSASSLGLGL | 22178444 |
| 23 | AINX_MOUSE | 54 | SRSNVASTAA C SSASSLGLGL | 22588120 |
| 24 | AINX_MOUSE | 54 | SRSNVASTAA C SSASSLGLGL | 24895380 |
| 25 | AINX_MOUSE | 54 | SRSNVASTAA C SSASSLGLGL | 19101475 |
| 26 | AL7A1_MOUSE | 478 | FRWLGPKGSD C GIVNVNIPTS | 22178444 |
| 27 | AL7A1_MOUSE | 478 | FRWLGPKGSD C GIVNVNIPTS | 21278135 |
| 28 | AL7A1_MOUSE | 478 | FRWLGPKGSD C GIVNVNIPTS | 20837516 |
| 29 | ALBU_MOUSE | 384 | AKKYEATLEK C CAEANPPACY | 22588120 |
| 30 | ALBU_MOUSE | 385 | KKYEATLEKC C AEANPPACYG | 22588120 |
| 31 | ALD2_MOUSE | 187 | KHKPVTNQVE C HPYLTQEKLI | 21278135 |
| 32 | ALDOA_MOUSE | 339 | IKRALANSLA C QGKYTPSGQS | 24895380 |
| 33 | ALDOA_MOUSE | 339 | IKRALANSLA C QGKYTPSGQS | 24926564 |
| 34 | ALDOA_MOUSE | 339 | IKRALANSLA C QGKYTPSGQS | 19101475 |
| 35 | ALDOA_MOUSE | 339 | IKRALANSLA C QGKYTPSGQS | 20925432 |
| 36 | ALDOA_MOUSE | 339 | IKRALANSLA C QGKYTPSGQS | 21278135 |
| 37 | ALDOA_MOUSE | 339 | IKRALANSLA C QGKYTPSGQS | 22865876 |
| 38 | ALDOB_MOUSE | 158 | WRAVLRIADQ C PSSLAIQENA | 20837516 |
| 39 | BAG3_MOUSE | 185 | ERSQSPAASD C SSSSSSASLP | 21278135 |
| 40 | BAG3_MOUSE | 185 | ERSQSPAASD C SSSSSSASLP | 22865876 |
| 41 | BCAT2_MOUSE | 229 | VRAWIGGVGD C KLGGNYGPTV | 21278135 |
| 42 | CA163_MOUSE | 111 | EKPGKKSVES C HNVGLLAHDG | 21278135 |
| 43 | CAH3_MOUSE | 66 | AKTILNNGKT C RVVFDDTYDR | 22178444 |
| 44 | CAH3_MOUSE | 66 | AKTILNNGKT C RVVFDDTYDR | 20837516 |
| 45 | CATS_MOUSE | 329 | IRMARNNKNH C GIASYCSYPE | 22178444 |
| 46 | CATS_MOUSE | 329 | IRMARNNKNH C GIASYCSYPE | 19483679 |
| 47 | CD81_MOUSE | 157 | VKTFHETLNC C GSNALTTLTT | 22588120 |
| 48 | CD81_MOUSE | 157 | VKTFHETLNC C GSNALTTLTT | 24895380 |
| 49 | CELF1_MOUSE | 150 | LRGPDGLSRG C AFVTFTTRTM | 20925432 |
| 50 | CK054_MOUSE | 226 | AHIMPAEFSS C PLNSDEAVNK | 22178444 |
| 51 | CK054_MOUSE | 226 | AHIMPAEFSS C PLNSDEAVNK | 20837516 |
| 52 | CPT1B_MOUSE | 526 | QRLPWDIPEQ C REAIENSYQV | 21278135 |
| 53 | CPT1B_MOUSE | 526 | QRLPWDIPEQ C REAIENSYQV | 22865876 |
| 54 | CPT2_MOUSE | 643 | AREFLHCVQK C LEDMFDALEG | 21278135 |
| 55 | CSN2_MOUSE | 179 | QKILRQLHQS C QTDDGEDDLK | 20925432 |
| 56 | CSRP3_MOUSE | 171 | DKDGELYCKV C YAKNFGPTGI | 21278135 |
| 57 | DCMC_MOUSE | 67 | EKTPAPAEGQ C ADFVSFYGGL | 22865876 |
| 58 | DCMC_MOUSE | 359 | GRNELFTDSE C QEISAVTGNP | 21278135 |
| 59 | DHB8_MOUSE | 192 | ARELGRHGIR C NSVLPGFIAT | 21278135 |
| 60 | DHE3_MOUSE | 327 | MRYLHRFGAK C VGVGESDGSI | 22588120 |
| 61 | DHE3_MOUSE | 327 | MRYLHRFGAK C VGVGESDGSI | 24895380 |
| 62 | DHE3_MOUSE | 327 | MRYLHRFGAK C VGVGESDGSI | 21278135 |
| 63 | DHE3_MOUSE | 327 | MRYLHRFGAK C VGVGESDGSI | 22865876 |
| 64 | DRG2_MOUSE | 270 | ARKPNSVVIS C GMKLNLDYLL | 21278135 |
| 65 | E9PUA9_MOUSE | 304 | ARFSYIPLAT C HFHINDLELS | 20925432 |
| 66 | ERI3_MOUSE | 285 | LKKAYSFAMG C WPKNGLLDMN | 21278135 |
| 67 | F6V8A7_MOUSE | 201 | NKMFNGSKLK C PYCPMEQSPG | 24926564 |
| 68 | FABP5_MOUSE | 43 | RKMAAMAKPD C IITCDGNNIT | 22178444 |
| 69 | FABP5_MOUSE | 43 | RKMAAMAKPD C IITCDGNNIT | 19483679 |
| 70 | FAS_MOUSE | 1464 | RKEPGGHRIR C ILLSNLSNTS | 23748021 |
| 71 | FAS_MOUSE | 1464 | RKEPGGHRIR C ILLSNLSNTS | 20837516 |
| 72 | FKB1A_MOUSE | 23 | GRTFPKRGQT C VVHYTGMLED | 21278135 |
| 73 | FKB1B_MOUSE | 23 | GRTFPKKGQI C VVHYTGMLQN | 21278135 |
| 74 | GCDH_MOUSE | 176 | PRLAKGELLG C FGLTEPNHGS | 21278135 |
| 75 | GRHPR_MOUSE | 29 | GRAALAQAAD C EVEQWNSDDP | 21278135 |
| 76 | GSTK1_MOUSE | 176 | NKLIENTDAA C KYGAFGLPTT | 21278135 |
| 77 | GSTK1_MOUSE | 176 | NKLIENTDAA C KYGAFGLPTT | 22865876 |
| 78 | GSTO1_MOUSE | 90 | GHLVTESVIT C EYLDEAYPEK | 21278135 |
| 79 | GYS1_MOUSE | 564 | DRRFRSLDDS C SQLTSFLYSF | 22865876 |
| 80 | H3BL49_MOUSE | 185 | VKDAKIAVYS C PFDGMITETK | 24926564 |
| 81 | HCDH_MOUSE | 211 | CKTLGKHPVS C KDTPGFIVNR | 21278135 |
| 82 | HCDH_MOUSE | 211 | CKTLGKHPVS C KDTPGFIVNR | 22865876 |
| 83 | HHATL_MOUSE | 326 | DHLDPPQPPK C ITALYVFGET | 22865876 |
| 84 | HKDC1_MOUSE | 438 | HKVVRRLVPN C DVRFLLSESG | 21278135 |
| 85 | HNRPU_MOUSE | 449 | EKPYFPIPED C TFIQNVPLED | 20925432 |
| 86 | HOP_MOUSE | 68 | WRRSEGLPSE C RSVTD | 21278135 |
| 87 | HOP_MOUSE | 68 | WRRSEGLPSE C RSVTD | 22865876 |
| 88 | IDHC_MOUSE | 269 | MKSEGGFIWA C KNYDGDVQSD | 24926564 |
| 89 | IDHC_MOUSE | 269 | MKSEGGFIWA C KNYDGDVQSD | 21278135 |
| 90 | IDHC_MOUSE | 269 | MKSEGGFIWA C KNYDGDVQSD | 22865876 |
| 91 | IDHP_MOUSE | 308 | LKSSGGFVWA C KNYDGDVQSD | 21278135 |
| 92 | IDHP_MOUSE | 308 | LKSSGGFVWA C KNYDGDVQSD | 22865876 |
| 93 | IDHP_MOUSE | 402 | IRFAQTLEKV C VQTVESGAMT | 21278135 |
| 94 | IDHP_MOUSE | 402 | IRFAQTLEKV C VQTVESGAMT | 22865876 |
| 95 | IF2A_MOUSE | 218 | VKEALRAGLN C STETMPIKIN | 20925432 |
| 96 | IF4G1_MOUSE | 940 | NHDEESLECL C RLLTTIGKDL | 24926564 |
| 97 | IPO5_MOUSE | 560 | LRLLRGKTIE C ISLIGLAVGK | 22865876 |
| 98 | IPYR_MOUSE | 242 | TKKTDGKGIS C MNTTVSESPF | 21278135 |
| 99 | IVD_MOUSE | 252 | DKLGMRGSNT C ELVFEDCKVP | 22865876 |
| 100 | KCNA2_MOUSE | 482 | FREENLKTAN C TLANTNYVNI | 24895380 |
| 101 | KGUA_MOUSE | 98 | VRAVQAMNRI C VLDVDLQGVR | 21278135 |
| 102 | KT3K_MOUSE | 24 | VKATGHSGGG C ISQGQSYDTD | 21278135 |
| 103 | LA_MOUSE | 18 | EKMTALEAKI C HQIEYYFGDF | 20925432 |
| 104 | LEG9_MOUSE | 73 | PRFEEGGYVV C NTKQNGQWGP | 20925432 |
| 105 | MACD1_MOUSE | 264 | EHRLRSVAFP C ISTGVFGYPN | 21278135 |
| 106 | MAP1B_MOUSE | 1913 | EKTERTIKSP C DSGYSYETIE | 22178444 |
| 107 | MAP1B_MOUSE | 1913 | EKTERTIKSP C DSGYSYETIE | 19101475 |
| 108 | MAP4_MOUSE | 595 | QHKGQSTVPP C TASPEPVKAA | 21278135 |
| 109 | MBB1A_MOUSE | 890 | MHHLCRARRY C HEVGPCAEAL | 20925432 |
| 110 | MDHM_MOUSE | 212 | GKTIIPLISQ C TPKVDFPQDQ | 22588120 |
| 111 | MDHM_MOUSE | 212 | GKTIIPLISQ C TPKVDFPQDQ | 24895380 |
| 112 | MDHM_MOUSE | 212 | GKTIIPLISQ C TPKVDFPQDQ | 21278135 |
| 113 | MDHM_MOUSE | 212 | GKTIIPLISQ C TPKVDFPQDQ | 22865876 |
| 114 | MECR_MOUSE | 345 | IRQGRLTAPS C SEVPLQGYQQ | 21278135 |
| 115 | MIRO1_MOUSE | 572 | HKMPPPQAFT C NTADAPSKDI | 21278135 |
| 116 | MMSA_MOUSE | 149 | FRGLQVVEHA C SVTSLMLGET | 21278135 |
| 117 | MMSA_MOUSE | 149 | FRGLQVVEHA C SVTSLMLGET | 22865876 |
| 118 | MPI_MOUSE | 29 | GKVGSKSEVA C LLASSDPLAQ | 21278135 |
| 119 | MTNB_MOUSE | 75 | ERIQPEDMFV C DINEQDISGP | 21278135 |
| 120 | MYH4_MOUSE | 699 | EHELVLHQLR C NGVLEGIRIC | 22865876 |
| 121 | MYH9_MOUSE | 91 | SKVEDMAELT C LNEASVLHNL | 20925432 |
| 122 | MYPC3_MOUSE | 784 | PKISNVGEDS C TVQWEPPAYD | 21278135 |
| 123 | MYPC3_MOUSE | 784 | PKISNVGEDS C TVQWEPPAYD | 22865876 |
| 124 | NADC_MOUSE | 111 | ERVALNTLAR C SGIASAAATA | 22178444 |
| 125 | NADC_MOUSE | 111 | ERVALNTLAR C SGIASAAATA | 20837516 |
| 126 | NCAM1_MOUSE | 139 | FKEGEDAVIV C DVVSSLPPTI | 24895380 |
| 127 | NDRG1_MOUSE | 289 | TKTTLLKMAD C GGLPQISQPA | 21278135 |
| 128 | NDUS2_MOUSE | 347 | MRQSLRIIEQ C LNKMPPGEIK | 22865876 |
| 129 | NDUV2_MOUSE | 223 | PKPGPRSGRF C CEPAGGLTSL | 24895380 |
| 130 | NDUV2_MOUSE | 223 | PKPGPRSGRF C CEPAGGLTSL | 22865876 |
| 131 | NFASC_MOUSE | 501 | IRKEDQGIYT C VATNILGKAE | 24895380 |
| 132 | NIBL1_MOUSE | 154 | SKSGSTPILK C PTQFPLILWH | 20925432 |
| 133 | NNRE_MOUSE | 109 | SKSPPTVLVI C GPGNNGGDGL | 22865876 |
| 134 | NTR2_MOUSE | 153 | ARRLLTPRRT C RLLSLVWVAS | 16418269 |
| 135 | ODPA_MOUSE | 94 | QKIIRGFCHL C DGQEACCVGL | 24895380 |
| 136 | ODPA_MOUSE | 94 | QKIIRGFCHL C DGQEACCVGL | 22865876 |
| 137 | ODPA_MOUSE | 101 | CHLCDGQEAC C VGLEAGINPT | 24895380 |
| 138 | ODPA_MOUSE | 101 | CHLCDGQEAC C VGLEAGINPT | 22865876 |
| 139 | OPA1_MOUSE | 874 | QRHFIDSELE C NDVVLFWRIQ | 22865876 |
| 140 | P5CS_MOUSE | 674 | LRTEYGDLEV C IEVVDSVQEA | 20925432 |
| 141 | PERM1_MOUSE | 807 | PRSSSSSNPS C | 22865876 |
| 142 | PFKAM_MOUSE | 1411 | KRGLVLRNEK C NENYTTDFIF | 22865876 |
| 143 | PPE2_MOUSE | 403 | RRSVDLELEQ C RQQAGFLGIR | 22865876 |
| 144 | PPP5_MOUSE | 77 | NRSLAYLRTE C YGYALGDATR | 22178444 |
| 145 | PPP5_MOUSE | 77 | NRSLAYLRTE C YGYALGDATR | 19101475 |
| 146 | PPP5_MOUSE | 77 | NRSLAYLRTE C YGYALGDATR | 21278135 |
| 147 | PPP5_MOUSE | 77 | NRSLAYLRTE C YGYALGDATR | 22865876 |
| 148 | PPP5_MOUSE | 404 | GRSVSKRGVS C QFGPDVTKAF | 21278135 |
| 149 | PRDX1_MOUSE | 71 | DRADEFKKLN C QVIGASVDSH | 22865876 |
| 150 | PRDX2_MOUSE | 70 | DHAEDFRKLG C EVLGVSVDSQ | 22865876 |
| 151 | PRDX3_MOUSE | 128 | DKANEFHDVN C EVVAVSVDSH | 22865876 |
| 152 | PRDX4_MOUSE | 151 | FKSINTEVVA C SVDSQFTHLA | 20925432 |
| 153 | PUR2_MOUSE | 134 | WRAFTNPEDA C SFITSANFPA | 21278135 |
| 154 | PURA1_MOUSE | 183 | SKAARTGLRI C DLLSDFDEFS | 21278135 |
| 155 | Q3TLP8_MOUSE | 124 | AKWYPEVRHH C PNTPIILVGT | 20925432 |
| 156 | Q3V117_MOUSE | 229 | AKVDATADYI C KVKWGDIEFP | 20925432 |
| 157 | RAC1_MOUSE | 105 | AKWYPEVRHH C PNTPIILVGT | 24895380 |
| 158 | RAC1_MOUSE | 105 | AKWYPEVRHH C PNTPIILVGT | 20925432 |
| 159 | RAC1_MOUSE | 105 | AKWYPEVRHH C PNTPIILVGT | 21278135 |
| 160 | RAC1_MOUSE | 105 | AKWYPEVRHH C PNTPIILVGT | 22865876 |
| 161 | RBM27_MOUSE | 49 | DKPEKELKAF C ADQLDVFLQK | 20925432 |
| 162 | RHOA_MOUSE | 107 | EKWTPEVKHF C PNVPIILVGN | 21278135 |
| 163 | RHOA_MOUSE | 107 | EKWTPEVKHF C PNVPIILVGN | 22865876 |
| 164 | RHOB_MOUSE | 107 | EKWVPEVKHF C PNVPIILVAN | 20925432 |
| 165 | RHOC_MOUSE | 107 | EKWTPEVKHF C PNVPIILVGN | 20925432 |
| 166 | RHOC_MOUSE | 107 | EKWTPEVKHF C PNVPIILVGN | 21278135 |
| 167 | RM13_MOUSE | 53 | NKPVYHQLSD C GDHVVIINTR | 21278135 |
| 168 | RS27A_MOUSE | 144 | SHFDRHYCGK C CLTYCFNKPE | 20925432 |
| 169 | RT07_MOUSE | 152 | YRIFHEALKN C EPVIGLVPIL | 21278135 |
| 170 | S61A1_MOUSE | 13 | IKFLEVIKPF C VILPEIQKPE | 22178444 |
| 171 | S61A1_MOUSE | 13 | IKFLEVIKPF C VILPEIQKPE | 20925432 |
| 172 | S61A1_MOUSE | 13 | IKFLEVIKPF C VILPEIQKPE | 20837516 |
| 173 | SFI1_MOUSE | 566 | HHSGLLRRAF C IWKESTQGFR | 21278135 |
| 174 | SMYD1_MOUSE | 412 | GHIEVGHGMI C KAYAILLVTH | 21278135 |
| 175 | SNAB_MOUSE | 103 | KKADPQEAIN C LNAAIDIYTD | 22178444 |
| 176 | SNAB_MOUSE | 103 | KKADPQEAIN C LNAAIDIYTD | 16418269 |
| 177 | SPP24_MOUSE | 109 | QRGYSVPTAA C RSTVQMSKGQ | 21278135 |
| 178 | SUV3_MOUSE | 418 | ARKFNDPNDP C KILVATDAIG | 21278135 |
| 179 | SYDC_MOUSE | 130 | VRKVNQKIGS C TQQDVELHVQ | 21278135 |
| 180 | SYEP_MOUSE | 744 | ERPAPAVSST C ATAEDSSVLY | 21278135 |
| 181 | SYVC_MOUSE | 478 | YRSTRLVNWS C TLNSAISDIE | 20925432 |
| 182 | TAGL2_MOUSE | 63 | QKWLKDGTVL C KLINSLYPEG | 21278135 |
| 183 | TB22A_MOUSE | 150 | VKSVSESHTP C PSESTGDTVP | 21278135 |
| 184 | TCPG_MOUSE | 398 | ERNLQDAMQV C RNVLLDPQLV | 21278135 |
| 185 | TCPQ_MOUSE | 244 | VKDAKIAVYS C PFDGMITETK | 24926564 |
| 186 | TGM2_MOUSE | 553 | LRILYEKYSG C LTESNLIKVR | 21278135 |
| 187 | TGM2_MOUSE | 553 | LRILYEKYSG C LTESNLIKVR | 22865876 |
| 188 | THIM_MOUSE | 287 | ARVVGYFVSG C DPTIMGIGPV | 22178444 |
| 189 | THIM_MOUSE | 287 | ARVVGYFVSG C DPTIMGIGPV | 23748021 |
| 190 | THIM_MOUSE | 287 | ARVVGYFVSG C DPTIMGIGPV | 21278135 |
| 191 | THIM_MOUSE | 287 | ARVVGYFVSG C DPTIMGIGPV | 20837516 |
| 192 | THIM_MOUSE | 287 | ARVVGYFVSG C DPTIMGIGPV | 22865876 |
| 193 | TITIN_MOUSE | 1730 | LKRFGPAHFE C RLTPIGDPTM | 22865876 |
| 194 | TITIN_MOUSE | 4321 | VKTEDEGEYV C EASNDSGKAK | 22865876 |
| 195 | TITIN_MOUSE | 16971 | GRYVITATNS C GSKFAAVRVE | 22865876 |
| 196 | TITIN_MOUSE | 17002 | LKPVVTNRKM C LLNWSDPADD | 22865876 |
| 197 | TITIN_MOUSE | 17066 | FRVIAKNKFG C GPPVEIGPIL | 22865876 |
| 198 | TITIN_MOUSE | 17754 | GKAWTKVNPN C GSTTFVVPDL | 22865876 |
| 199 | TITIN_MOUSE | 20504 | LKVSDVTKTS C HVSWAPPEND | 22865876 |
| 200 | TITIN_MOUSE | 26608 | LKVTGVTAEK C YLAWNPPLQD | 22865876 |
| 201 | TITIN_MOUSE | 29692 | KRQGSDRWVR C NFTDVSECQY | 22865876 |
| 202 | TITIN_MOUSE | 29700 | VRCNFTDVSE C QYTVTGLSPG | 22865876 |
| 203 | TITIN_MOUSE | 34371 | VHEGESARFS C DTDGEPVPTV | 22865876 |
| 204 | TLN1_MOUSE | 1199 | AKAVTQALNR C VSCLPGQRDV | 20925432 |
| 205 | TPP2_MOUSE | 614 | LREGLHYTEV C GYDIASPNAG | 20925432 |
| 206 | TRAP1_MOUSE | 263 | TKIIIHLKSD C KDFASESRVQ | 22178444 |
| 207 | TRAP1_MOUSE | 263 | TKIIIHLKSD C KDFASESRVQ | 21278135 |
| 208 | TRAP1_MOUSE | 263 | TKIIIHLKSD C KDFASESRVQ | 20837516 |
| 209 | UB2V1_MOUSE | 144 | MKLPQPPEGQ C YSN | 21278135 |
| 210 | UBL5_MOUSE | 18 | DRLGKKVRVK C NTDDTIGDLK | 20925432 |
| 211 | UBL5_MOUSE | 18 | DRLGKKVRVK C NTDDTIGDLK | 21278135 |
| 212 | UCP3_MOUSE | 25 | VKFLGAGTAA C FADLLTFPLD | 21278135 |
| 213 | VLDLR_MOUSE | 120 | MRTCRINEIS C GARSTQCIPV | 21278135 |
| 214 | WDR1_MOUSE | 170 | YRLATGSDDN C AAFFEGPPFK | 21278135 |
| 215 | ZADH2_MOUSE | 55 | FHEAVTLRRD C PVPLPGDGDL | 21278135 |
